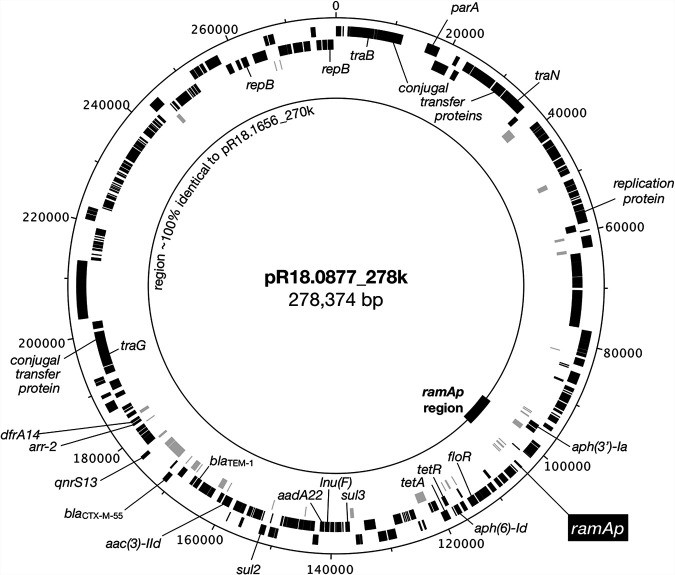
The ramA gene plays a crucial role in antimicrobial resistance in bacteria. This gene encodes the RamA protein, a transcriptional regulator that controls the expression of efflux pumps. Efflux pumps are responsible for removing harmful substances, including antibiotics, from bacterial cells, contributing to multidrug resistance. This article explores a plasmid-borne ramA gene, ramAp, found in Salmonella enterica serovar Goldcoast and its impact on antibiotic susceptibility.
RamA: A Key Regulator of Efflux Pumps
Bacteria utilize efflux pumps to expel various compounds, including antibiotics, from their cells. The AcrAB-TolC efflux pump, prevalent in Escherichia coli and Salmonella, expels drugs like quinolones, chloramphenicol, and tetracycline. Overexpression of this pump significantly reduces bacterial susceptibility to these antibiotics. The ramA gene encodes the RamA regulator protein, which directly binds to and activates the promoters of acrAB and tolC, leading to their increased expression and subsequent drug resistance.
Plasmid-Borne ramA: A New Resistance Determinant
Researchers have identified a plasmid-borne ramA gene, ramAp, in a multidrug-resistant Salmonella enterica serovar Goldcoast strain. This gene is located on a large IncHI2 plasmid and is flanked by insertion sequence elements, suggesting its potential for mobilization and transfer between bacterial species. Comparative genomic analysis revealed that ramAp is identical to the chromosomal ramA gene found in Klebsiella pneumoniae.
Figure 1: Comparison of plasmids pR18.0877_278k and pR18.1656_270k highlighting the ramAp region. Plasmid pR18.0877_278k harbors an extra 6.6-kb region containing the ramAp gene, absent in pR18.1656_270k. This region is characterized by multiple insertion sequence elements and antimicrobial resistance genes.
RamAp Increases Antibiotic Resistance
Introducing ramAp into E. coli significantly increased resistance to various antibiotics, including chloramphenicol, azithromycin, nalidixic acid, ciprofloxacin, and tetracycline. This heightened resistance was reversed by the efflux pump inhibitor phenylalanine-arginine β-naphthylamide (PaβN), confirming the role of RamAp in activating efflux pumps.
Figure 2: The 6.6-kb ramAp region in plasmid pR18.0877_278k. The ramAp gene is flanked by a truncated ISEcp1 element and other genetic features. The region shows high similarity to the chromosomal ramA loci of Klebsiella quasipneumoniae.
Quantitative PCR analysis demonstrated that ramAp expression led to a significant increase in the expression of acrA, acrB, and tolC genes, further supporting its role in enhancing efflux pump activity. This upregulation was observed in both E. coli and Salmonella strains harboring ramAp.
Figure 3: Expression levels of efflux pump genes in E. coli and Salmonella strains. ramAp significantly increases the expression of acrA, acrB, and tolC, leading to enhanced efflux pump activity and antibiotic resistance.
Implications for Public Health
The discovery of a plasmid-borne ramA gene raises concerns about the spread of antibiotic resistance. The IncHI2 plasmid carrying ramAp can potentially transfer this resistance determinant to other bacterial species, contributing to the global challenge of multidrug-resistant infections. Further research is crucial to understand the prevalence and dissemination of ramAp and to develop strategies to mitigate its impact on public health.
Conclusion
The ramAp gene represents a significant threat to antibiotic effectiveness. Its ability to enhance efflux pump activity and confer resistance to multiple antibiotics highlights the importance of monitoring its spread and developing novel therapeutic approaches to combat this emerging resistance mechanism.